1. GBD 2017 Inflammatory Bowel Disease Collaborators. The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol 2020;5:17-30.

2. Maaser C, Sturm A, Vavricka SR, et al. ECCO-ESGAR Guideline for Diagnostic Assessment in IBD Part 1: initial diagnosis, monitoring of known IBD, detection of complications. J Crohns Colitis 2019;13:144-164.


4. Gajendran M, Loganathan P, Catinella AP, Hashash JG. A comprehensive review and update on Crohn’s disease. Dis Mon 2018;64:20-57.


5. Gajendran M, Loganathan P, Jimenez G, et al. A comprehensive review and update on ulcerative colitis. Dis Mon 2019;65:100851.


6. Dal Buono A, Roda G, Argollo M, Peyrin-Biroulet L, Danese S. Histological healing: should it be considered as a new outcome for ulcerative colitis? Expert Opin Biol Ther 2020;20:407-412.


7. Turner D, Ricciuto A, Lewis A, et al. STRIDE-II: an update on the selecting therapeutic targets in inflammatory bowel disease (STRIDE) initiative of the International Organization for the Study of IBD (IOIBD): determining therapeutic goals for treat-to-target strategies in IBD. Gastroenterology 2021;160:1570-1583.


8. Schett G, McInnes IB, Neurath MF. Reframing immune-mediated inflammatory diseases through signature cytokine hubs. N Engl J Med 2021;385:628-639.


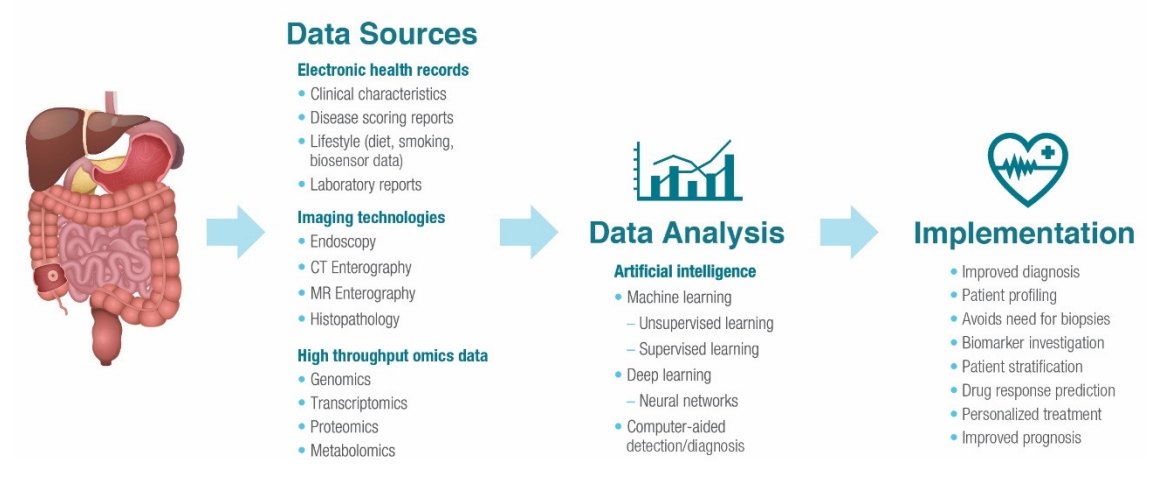
9. Le Berre C, Sandborn WJ, Aridhi S, et al. Application of artificial intelligence to gastroenterology and hepatology. Gastroenterology 2020;158:76-94.


12. Chahal D, Byrne MF. A primer on artificial intelligence and its application to endoscopy. Gastrointest Endosc 2020;92:813-820.


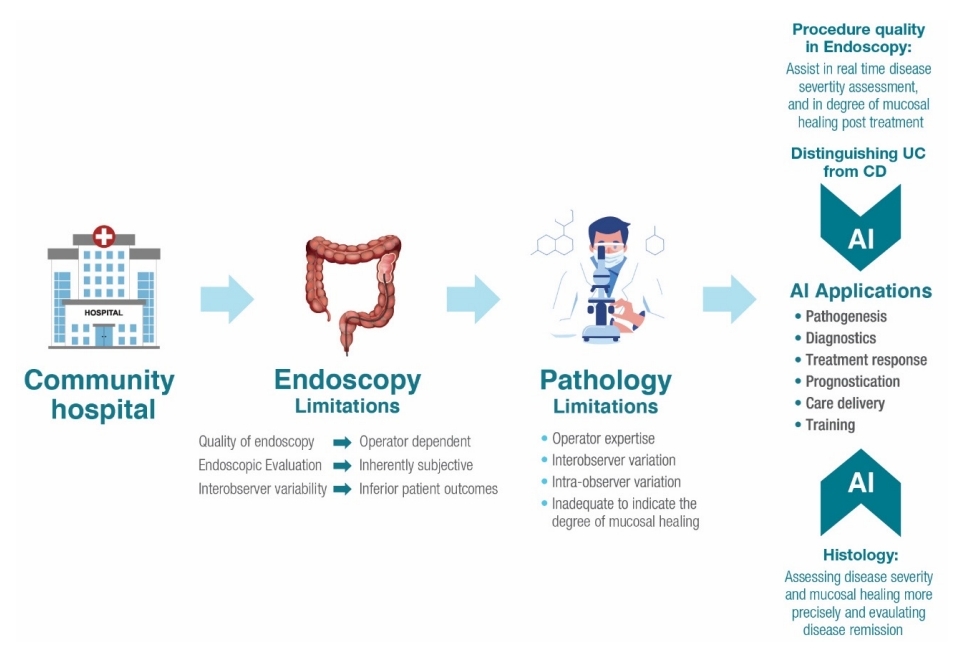
14. Fernandes SR, Pinto JS, Marques da Costa P, Correia L, GEDII. Disagreement among gastroenterologists using the Mayo and Rutgeerts Endoscopic Scores. Inflamm Bowel Dis 2018;24:254-260.


15. Stidham R, Yao H, Soroushmehr R, et al. 796 Computer vision measurement of disease severity distribution outperforms traditional endoscopic scoring for detecting therapeutic response in a clinical trial of ustekinumab for ulcerative colitis. Gastroenterology 2022;162:S-193.

16. Maeda Y, Kudo SE, Mori Y, et al. Fully automated diagnostic system with artificial intelligence using endocytoscopy to identify the presence of histologic inflammation associated with ulcerative colitis (with video). Gastrointest Endosc 2019;89:408-415.


19. Berzin TM, Parasa S, Wallace MB, Gross SA, Repici A, Sharma P. Position statement on priorities for artificial intelligence in GI endoscopy: a report by the ASGE Task Force. Gastrointest Endosc 2020;92:951-959.


20. Dekker E, Nass KJ, Iacucci M, et al. Performance measures for colonoscopy in inflammatory bowel disease patients: European Society of Gastrointestinal Endoscopy (ESGE) Quality Improvement Initiative. Endoscopy 2022;54:904-915.


22. Freedman D, Blau Y, Katzir L, et al. Detecting deficient coverage in colonoscopies. IEEE Trans Med Imaging 2020;39:3451-3462.


23. Zhou J, Wu L, Wan X, et al. A novel artificial intelligence system for the assessment of bowel preparation (with video). Gastrointest Endosc 2020;91:428-435.


25. Ali S, Zhou F, Bailey A, et al. A deep learning framework for quality assessment and restoration in video endoscopy. Med Image Anal 2021;68:101900.


27. Bielecki C, Bocklitz TW, Schmitt M, et al. Classification of inflammatory bowel diseases by means of Raman spectroscopic imaging of epithelium cells. J Biomed Opt 2012;17:076030.


30. Huang TY, Zhan SQ, Chen PJ, Yang CW, Lu HH. Accurate diagnosis of endoscopic mucosal healing in ulcerative colitis using deep learning and machine learning. J Chin Med Assoc 2021;84:678-681.


31. Yao H, Najarian K, Gryak J, et al. Fully automated endoscopic disease activity assessment in ulcerative colitis. Gastrointest Endosc 2021;93:728-736.


32. Gutierrez Becker B, Arcadu F, Thalhammer A, et al. Training and deploying a deep learning model for endoscopic severity grading in ulcerative colitis using multicenter clinical trial data. Ther Adv Gastrointest Endosc 2021;14:2631774521990623.


33. Takenaka K, Ohtsuka K, Fujii T, et al. Development and validation of a deep neural network for accurate evaluation of endoscopic images from patients with ulcerative colitis. Gastroenterology 2020;158:2150-2157.


34. Bossuyt P, Nakase H, Vermeire S, et al. Automatic, computer-aided determination of endoscopic and histological inflammation in patients with mild to moderate ulcerative colitis based on red density. Gut 2020;69:1778-1786.


41. Guidi L, Marzo M, Andrisani G, et al. Faecal calprotectin assay after induction with anti-tumour necrosis factor α agents in inflammatory bowel disease: prediction of clinical response and mucosal healing at one year. Dig Liver Dis 2014;46:974-979.


44. Verstockt B, Sudahakar P, Creyns B, et al. DOP70 An integrated multi-omics biomarker predicting endoscopic response in ustekinumab treated patients with Crohn’s disease. J Crohns Colitis 2019;13(Supplement_1):S072-S073.

47. D’Haens G, Kelly O, Battat R, et al. Development and validation of a test to monitor endoscopic activity in patients with Crohn’s disease based on serum levels of proteins. Gastroenterology 2020;158:515-526.


48. Sossenheimer PH, Yvellez OV, Andersen MJ, et al. 539 Wearable devices can predict disease activity in inflammatory bowel disease patients. Gastroenterology 2019;156:S-111.

53. Cheng JY, Abel JT, Balis UG, McClintock DS, Pantanowitz L. Challenges in the development, deployment, and regulation of artificial intelligence in anatomic pathology. Am J Pathol 2021;191:1684-1692.


54. Ehteshami Bejnordi B, Veta M, Johannes van Diest P, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 2017;318:2199-2210.


55. Saha M, Chakraborty C, Racoceanu D. Efficient deep learning model for mitosis detection using breast histopathology images. Comput Med Imaging Graph 2018;64:29-40.


57. Raya-Povedano JL, Romero-Martín S, Elías-Cabot E, Gubern-Mérida A, Rodríguez-Ruiz A, Álvarez-Benito M. AI-based strategies to reduce workload in breast cancer screening with mammography and tomosynthesis: a retrospective evaluation. Radiology 2021;300:57-65.


58. de Groof AJ, Struyvenberg MR, van der Putten J, et al. Deeplearning system detects neoplasia in patients with Barrett’s esophagus with higher accuracy than endoscopists in a multistep training and validation study with benchmarking. Gastroenterology 2020;158:915-929.


59. Luo H, Xu G, Li C, et al. Real-time artificial intelligence for detection of upper gastrointestinal cancer by endoscopy: a multicentre, case-control, diagnostic study. Lancet Oncol 2019;20:1645-1654.


60. Ebigbo A, Mendel R, Rückert T, et al. Endoscopic prediction of submucosal invasion in Barrett’s cancer with the use of artificial intelligence: a pilot study. Endoscopy 2021;53:878-883.


61. Kudo SE, Misawa M, Mori Y, et al. Artificial intelligence-assisted system improves endoscopic identification of colorectal neoplasms. Clin Gastroenterol Hepatol 2020;18:1874-1881.


65. Walradt T, Glissen Brown JR, Alagappan M, Lerner HP, Berzin TM. Regulatory considerations for artificial intelligence technologies in GI endoscopy. Gastrointest Endosc 2020;92:801-806.


68. Mori Y, Kudo SE, East JE, et al. Cost savings in colonoscopy with artificial intelligence-aided polyp diagnosis: an add-on analysis of a clinical trial (with video). Gastrointest Endosc 2020;92:905-911.


70. Rex DK. Making a resect-and-discard strategy work for diminutive colorectal polyps: let’s get real. Endoscopy 2022;54:364-366.