Precision medicine in inflammatory bowel diseases
Article information
Abstract
Inflammatory bowel diseases comprising Crohn’s disease and ulcerative colitis have emerged as global diseases. Multiple distinct therapeutic mechanisms have allowed us to increase our rates of achieving remission and reducing permanent disease-related morbidity. However, there is limited data to inform relative positioning of different therapies. This review will summarize existing literature on use of clinical decision models to inform relative efficacy of one therapeutic mechanism compared to the other given individual patient characteristics. It will also demonstrate the value of serologic, transcriptomic (from biopsies), and microbiome-based biomarkers in identifying which therapy is most likely to work for a given patient. We will review the existing gaps in the literature in this field and suggest a path forward for future studies to better inform patient care, incorporating the principles of precision medicine in the management of inflammatory bowel disease.
INTRODUCTION
Crohn’s disease (CD) and ulcerative colitis (UC), together termed inflammatory bowel diseases (IBD) have emerged as global diseases over with rising incidence in nearly every region of the world [1]. They are chronic, immune-mediated diseases that frequently require long-term treatment to reduce disease-related morbidity. The past two decades have witnessed the emergence of many effective therapies for these diseases that have improved our ability to achieve remission [2,3]. Three monoclonal antibodies against tumor necrosis factor α (anti-TNF) each are approved in the United States for CD and UC. In addition, 2 anti-integrins, and one antibody each against interleukin (IL)-12/23 and IL-23 are also approved for these conditions with others demonstrating promise in late-stage clinical trials. In addition to conventional immunomodulators, oral treatments have expanded to include both an S1P (sphingosine-1-phosphate) modulator and 2 Janus-kinase inhibitors [4]. However, despite the advances that have made possible these multiple different mechanisms of action, there is limited data to guide selection of treatment for a particular patient. Treatment choice is often dictated by factors other than likelihood of biologic response for a given individual patient. Consequently, both in observational data and in randomized controlled trials, there appears to be a therapeutic ceiling with a sizeable majority of patients failing to respond durably to any given therapeutic class [5]. The ability to a priori predict likelihood of response to maximize benefit for a given patient underlies the strategy of “precision medicine” in the management of IBD. Precision medicine is defined as medical care designed to optimize therapeutic benefit in particular groups of patients, often by molecular profiling. In clinical medicine, there are numerous examples of this ranging from assessing blood-group compatibility prior to transfusion to sophisticated molecular phenotyping of tumors to guide therapeutic regimen. This article will review some of the clinical and biomarker-based tools that have been proposed to inform therapeutic positioning for an individual patient (Fig. 1).
TOOLS FOR PROGNOSIS
There have been considerable strides towards identifying factors determining prognosis in CD and UC. Most tools have been developed to predict likelihood of colectomy in UC or need penetrating or stricturing complications or surgery in CD. In both diseases, a younger age of diagnosis is a predictor of a more unfavorable disease course. In CD, need for steroids at diagnosis, upper gastrointestinal involvement, perianal disease, ileocolonic disease location (compared to colon-only disease) and stricturing or penetrating behavior predicts greater likelihood of surgery and disabling course [6,7]. In UC, female sex, extensive colitis, low serum albumin, and elevated inflammatory markers predict a more aggressive disease course. Serologic or genetic markers may also predict a more aggressive course of CD. Siegel et al. [8] identified a cohort of 343 adult patients with CD among whom 142 progressed to stricturing or penetrating disease or required CD-related surgery. Anti-Saccharomyces cerevisiae antibodies and CBir1 as well as nucleotide-binding oligomerization domain-containing protein 2 (NOD2) mutations were independently associated with developing the adverse study outcome. In UC, αVβ6 autoantibodies have been associated with both risk of development of disease as well as complicated disease course [9]. In a cohort of newly diagnosed children with CD, baseline microbial composition also predicted disease progression. Greater abundance of Veillonella in the stool was associated with increased risk of penetrating disease while Ruminococcus abundance predicted stricturing complications. Extracellular matrix regulated ileal gene signatures were also associated with worse course of CD. In children with newly diagnosed UC, a model that incorporated hemoglobin at diagnosis, early treatment response at week 4, an antimicrobial peptide gene signature as well as abundance of 2 bacterial species (Ruminococcaceae and Sutterella) was predictive of disease course at 1 year with an area under the curve (AUC) of 0.87. Other studies have applied transcriptomics in tissue or whole blood to predict disease course. In a U.K. cohort of patients with CD, Biasci et al. [10] identified 2 clusters of transcripts on CD8+ T cells termed IBD1 and IBD2 subgroups. Patients belonging to IBD1 had a more aggressive disease course and were more refractory to therapy with a sensitivity of 73% in CD and 100% in UC.
PREDICTION OF THERAPY RESPONSE
1. Clinical Models
Multiple studies have examined predictors of response to various treatments in CD and UC [11,12]. Earlier initiation of treatment is predictive of better response while more severe disease (clinical symptoms, elevated laboratory markers or endoscopic severity) predicts lower rates of response. Attainment of higher serum trough biologic concentration, particularly with antiTNF agents, and use of combination immunomodulator treatment are also associated with higher rates of response in both CD and UC. Patients with prior biologic exposure have lower rates of response to subsequent therapy than biologic naïve patients [13,14]. Studies have also utilized such parameters to develop clinical decision support tools for implementation. For example, Vande Casteele et al. [15] developed a prediction model using sex, albumin, stool frequency, and rectal bleeding to identify likelihood of response to infliximab treatment. While these individual decision models are useful, many of the predictors are common to multiple therapeutic mechanisms, limiting their value in selecting treatment. However, there may be value to such models in optimizing response rates. For example, those determined a prior to having a lower likelihood of response may improve their outcomes through strategies such as a higher dose, use of combination therapy and proactive therapeutic monitoring post-induction. The efficacy of such approaches remains to be robustly established and merit further study.
Other studies have used clinical data to inform relative efficacy of one mechanism over another for a particular patient. Dulai et al. [16] used data from the GEMINI 2 trial of vedolizumab in CD to develop a model for prediction of response to treatment at 26 weeks. In a cohort of 814 patients, the absence of prior bowel surgery (+2 points), no prior anti-TNF use (+3 points), no prior fistulizing disease (+2 points), serum albumin (+0.4 points for each 1 g/dL) and baseline C-reactive protein grouped together in a score classified patients as low, intermediate or high likelihood of response to vedolizumab. A similar model in UC identified absence of prior anti-TNF exposure (+3 points), disease duration of 2 years or more (+3 points), baseline endoscopic severity (+2 points), and baseline albumin concentration (+0.65 per 1 g/dL) to predict response to vedolizumab treatment [17]. When applying the model to the VARSITY trial of adalimumab compared to vedolizumab in UC, the model was able to demonstrate a higher rate of week 52 clinical remission (39.7% vs. 24.2%) and histo-endoscopic mucosal healing (33.6% vs. 16.1%) with vedolizumab compared to adalimumab in those deemed to have high likelihood of response to vedolizumab using the decision support tool [18]. Similar decision support tools have also been developed for ustekinumab use incorporating prior anti-TNF exposure, bowel surgery, smoking status, albumin, and fistulizing disease at baseline. The ustekinumab decision support tool developing using data from the UNITI trials was applied to a Korean real-world cohort, demonstrating ability to differentiate low-probability and high-probability responders [19]. Narula et al. [20] used post-doc data from UNITI, VERSIFY, CT-P13, and EXTEND trials to demonstrate that in patients with ileal disease with large ulcers, both infliximab and adalimumab had higher rates of clinical benefit compared to ustekinumab or vedolizumab.
Clinical parameters may also inform choice of treatment from the standpoint of safety. In 2 large administrative claimsbased studies the likelihood of infectious complications was lower with vedolizumab compared to anti-TNF treatment among those with UC [21,22]. In a cohort that included 2,369 anti-TNF, 972 vedolizumab, and 352 ustekinumab users, among those with significant comorbidity defined as a Charlson Comorbidity Index > 1, vedolizumab (odds ratio, 0.78; 95% confidence interval, 0.65–0.94) and ustekinumab (odds ratio, 0.66; 95% confidence interval, 0.46–0.91) were both associated with lower rates of infection than anti-TNF treatment [23].
2. Molecular Predictors
1) Genetics
Over 250 distinct single nucleotide polymorphisms have been associated with CD or UC. However, apart from the association between NOD2 variants and development of stricturing or penetrating small-bowel CD-related complications, few variants have robustly and consistently been associated with disease phenotype including therapy response. While individual studies have identified single gene variants or used polygenic risk scores to predict response to anti-TNF therapy, these have not been replicated in independent cohorts [24,25].
2) Serum Biomarkers and Transcriptomics
Several studies have examined serologic markers or gene expression in tissue to identify if specific expression patterns predict response or nonresponse. Arijs et al. [26] used microarray data applied to colonic biopsies from 2 cohorts of patients with UC receiving treatment with infliximab. Between the 2 cohorts, there were 53 known genes that were differentially expressed at baseline between responders and non-responders to treatment. The top 5 differentially expressed genes were osteoprotegerin (TNFRSF11B), stanniocalcin-1 (STC1), prostaglandinendoperoxide synthase 2 (PTGS2), interleukin-13 receptor alpha 2 (IL-13Rα2), and IL-11. While applying a similar approach, gene expression data had a more modest predictive value in ileal CD [27]. West et al. [28] applied a sequential approach to transcriptomic data to identify signatures of treatment response. When comparing healthy control and IBD intestinal tissue, they identified 16 genes that were differentially expressed, of which 4 genes–oncostatin M (OSM), IL-1A, IL-1B, and IL-6 demonstrated at least a 2-fold difference and were common to both CD and UC. Expression of both OSM and OSM receptors demonstrated a dose-dependent relationship, progressively increasing with severity of disease on endoscopy and histopathology. Nearly 90% of patients who belonged to a cluster with high OSM expression at baseline were refractory to infliximab compared to only 15% of those who belonged to a low expression cluster. This association was independently validated in 2 independent clinical trial cohorts. Verstockt et al. [29] found whole blood triggering receptor expressed on myeloid cells 1 (TREM1) expression as well as mucosal TREM1 expression to be downregulated in patients who were likely to respond to anti-TNF therapy in both CD and UC with an AUC of ~0.77 to 0.78. This expression was specific to anti-TNF therapy as neither TREM1 expression nor serum TREM1 levels were differential between responders and non-responders to vedolizumab or ustekinumab. The same group also performed RNA-seq on colon biopsies of patients treated with vedolizumab or anti-TNF therapy and found baseline expression of 4 genes (PIWIL1, MAATS1, RGS13, and DCHS2) to predict endoscopic remission with vedolizumab (with an AUC of 0.79) but not anti-TNF treatment [30].
Deeper profiling at the single-cell transcriptional level has also been informative in predicting response to treatment in individual studies. Martin and colleagues applied single-cell transcriptomics to ileal biopsies of patients with CD [31]. They found a cellular module consisting of IgG plasma cells, inflammatory mononuclear phagocytes, activated T cells, and stromal cells (termed GIMATS) to be associated with likelihood of response to anti-TNF treatment. Patients who were resistant to anti-TNF treatment were enriched in higher expression of the GIMATS module (P=0.02) [31]. Similarly, Smillie et al. [32] demonstrated that in UC, the presence of IL-13Rα2+IL-11+inflammatory fibroblasts was associated with resistance to anti-TNF treatment.
3) Microbiome
The gut microbiome is central to the pathogenesis of IBD. Thus, it is plausible that microbial composition at baseline influences natural history of disease including response to treatment. There have been several studies that have demonstrated the gut microbiome to predict response to individual therapeutic mechanisms with fewer studies that have looked across mechanisms. In one of the earliest studies examining this association, Kolho et al. [33] demonstrated that microbial diversity at baseline or similarity to healthy controls predicted fecal calprotectin levels 3 months after initiation of anti-TNF treatment. In a study by Sanchis-Artero et al. [34], a ratio of Faecalibacterium to Escherichia coli was predictive of response to treatment (AUC, 0.87), with a higher accuracy than for calprotectin or symptom-based disease activity scores. In a Chinese cohort of 16 patients who were treated with infliximab and followed for 30 weeks, Zhou et al. [35] found several species to be differential between responders and non-responders to treatment. A model that was trained on both microbial composition using 16s rRNA sequencing and fecal calprotectin levels had the highest accuracy in predicting response to infliximab (AUC, 0.92). In a prospective cohort of patients initiating vedolizumab treatment at a single center, we demonstrated that abundance of 2 butyrate producing species, Burkholderiales and Roseburia inulinivorans was differential between responders and non-responders to treatment [36]. Importantly, at a functional level there were several pathways (13 in CD, 5 in UC) that were differentially distributed at baseline between responders and non-responders to treatment. An artificial neural network-based model that incorporated both compositional and functional changes in the microbiome predicted early treatment response with an AUC of 0.87. We then extended this study to include an additional independent cohort of 100 patients initiating anti-cytokine treatment (either anti-TNF or anti-IL-12/23) [37]. At baseline, the microbial composition could be grouped into 2 distinct clusters. Among patients who were assigned to cluster 1, the response rates to anti-TNF therapy at week 14 (72%) or week 52 (65%) were much higher than for anti-integrin treatment (41% and 26%) respectively. No difference was observed between the 2 treatment arms for those belonging to cluster 2. The higher likelihood of response to anti-TNF therapy was associated with higher serum secondary bile acid concentrations at baseline which, in turn, was linked, to a higher Bai+ operon positivity in patients whose microbiome resembled cluster 1.
PREDICTING SAFETY AND DOSING
Beyond just selection of the right treatment, precision medicine may also play a role in predicting durability to treatment. In the PANTS study of 1,240 biologic naïve-CD patients initiating infliximab or adalimumab, carriage of at least 1 HLA-DQA1* 05 allele was associated with likelihood of antibody formation to anti-TNF treatment, resulting in loss of response [38]. Ninety percent of HLA-DQA1*05 carriers on infliximab monotherapy developed antibodies to treatment at 1 year compared to only 25% of non-carrier on adalimumab combination therapy with an immunomodulator. Genetic polymorphisms in TPMT (thiopurine methyltransferase) and NUDT15 (Nudix hydrolase 15) are associated with leukopenia with thiopurine therapy [39] while HLA-DQA1*02:01/DRB1*07:01 haplotype is associated with an increase in risk of thiopurine-induced pancreatitis [40]. Precision medicine approaches can also be applied to deliver the right dose of medications. Such dashboard-guided treatment strategy that incorporates individual patient characteristics including drug clearance has demonstrated comparable or superior outcome when compared to conventional treatment while allowing for dose de-escalation in a substantial number of patients [41,42].
LIMITATIONS AND FUTURE DIRECTIONS
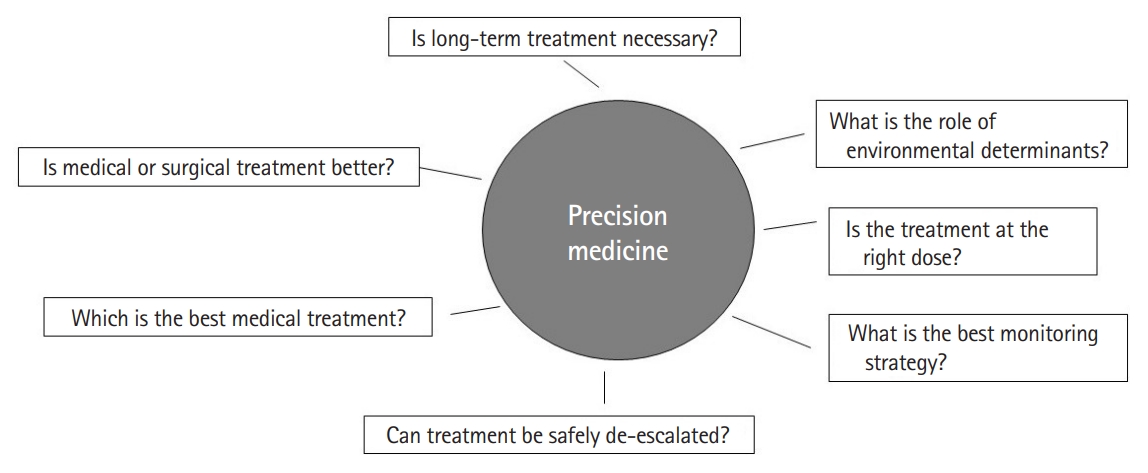
While the existing literature on the possibility of a precision medicine approach in IBD is promising, there are a number of limitations that need to be addressed in future studies. Importantly, studies have been heterogeneous in their methods including definitions of outcomes (such as symptomatic treatment response vs. endoscopic improvement), tools used to profile biomarkers, and adjustments for relevant confounders. Consequently, there have been few successful attempts at independently validating proposed biomarkers. There is an important need for a collaborative and synchronized multicenter effort to robustly define predictive biomarkers in IBD using pre-treatment samples and simultaneous endoscopic, imaging, and histologic evaluation with outcomes defined using standardized criteria. Second, most studies have been in Western populations. Emerging literature suggests that while there are common mechanisms, there are also key distinctions in the biological architecture of IBD between those of European ancestry and non-European ancestry cohorts. Thus, there is an important need to expand study in diverse populations. There is also a need for prospective interventional studies comparing treatment strategies based on biomarkers and predictive models. In the selection of treatments, as part of precision medicine, it is also important to involve patient preferences. This is important to set treatment goals and priorities as well as incorporate factors such as preferred route of administration and relative weights placed on efficacy and safety for each individual patient. In the selection of treatments, as part of precision medicine, it is also important to involve patient preferences. This is important to set treatment goals and priorities as well as incorporate factors such as preferred route of administration and relative weights placed on efficacy and safety for each individual patient. In the selection of treatments, as part of precision medicine, it is also important to involve patient preferences. This is important to set treatment goals and priorities as well as incorporate factors such as preferred route of administration and relative weights placed on efficacy and safety for each individual patient. In the selection of treatments, as part of precision medicine, it is also important to involve patient preferences. This is important to set treatment goals and priorities as well as incorporate factors such as preferred route of administration and relative weights placed on efficacy and safety for each individual patient. Similar to variability in response to medical therapy, there is also heterogeneity in which environmental factor(s) may be relevant in influencing treatment outcomes or risk of relapse in each patient. The goal of precision medicine is not just identifying the right medical treatment for the patient, but a broader aim of identifying which treatment option may be the best for any given patient (medical, surgical, dietary, and lifestyle), who may not need long-term treatment, and which patients may be appropriate candidates for de-escalation or early proactive escalated treatment (Fig. 2). Answering such questions requires international collaborative consortia and harmonization of efforts to arrive at robust solutions to improve our patient outcomes.
Notes
Funding Source
Ananthakrishnan AN was supported in part by grants from the National Institutes of Health (R21 DK127227, R01-DK127171) and the Chleck Family Foundation.
Conflict of Interest
No potential conflict of interest relevant to this article was reported.
Data Availability Statement
Not applicable.
Author Contribution
Writing and approval of the final manuscript: Ananthakrishnan AN.